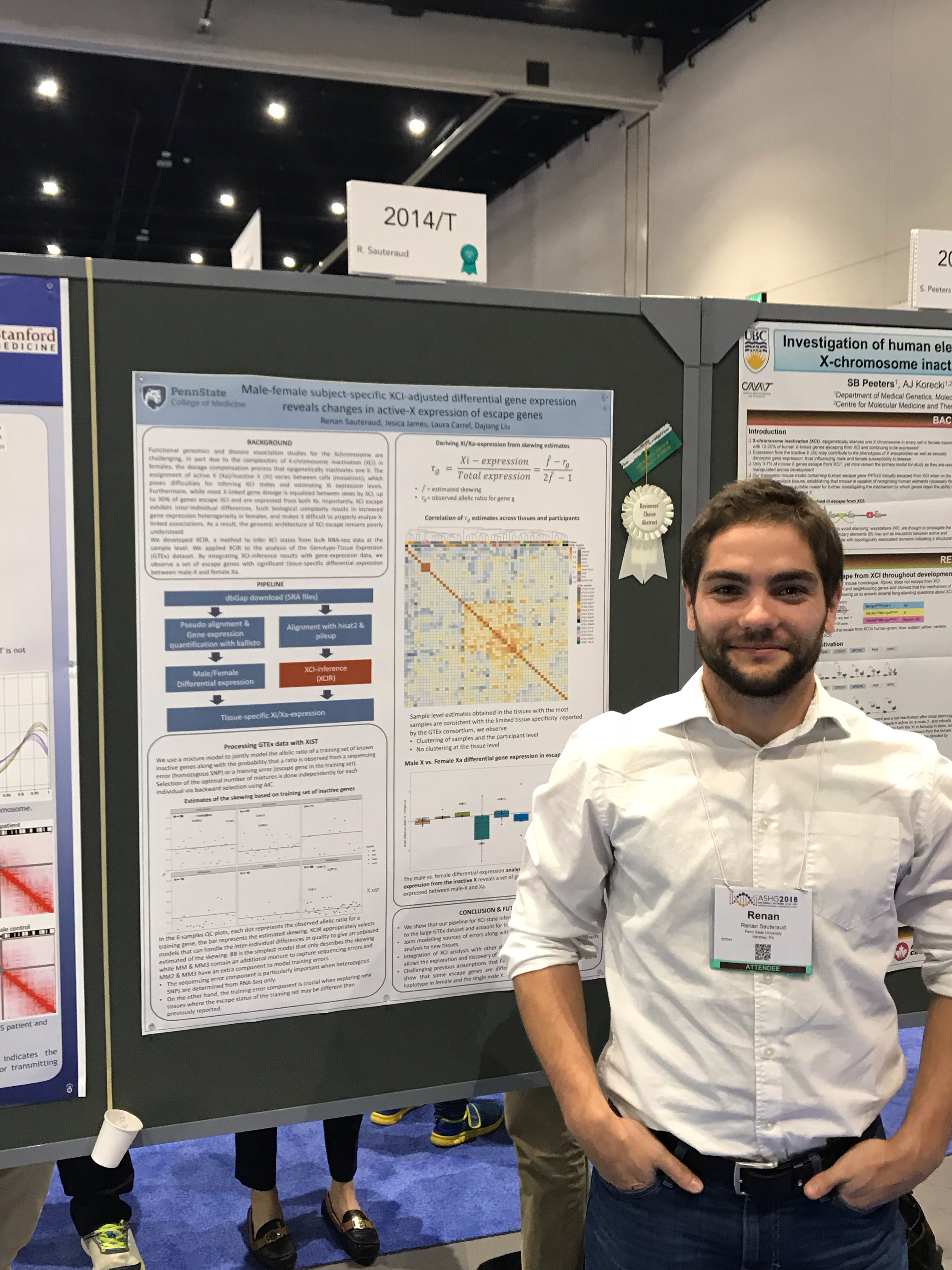
I am a third year Biostatistics PhD student at Penn State University. My current research topic is the inactivation of the X-chromosome (XCI) in females and the associated escape mechanisms. With my PhD advisor, Dr Dajiang Liu, I develop statistical models and software for the prediction of XCI status and study its association with various X-linked diseases and gender biased phenotypes.
Until 2016, I worked as a scientific programmer at the Fred Hutchinson Cancer Research Center in the RGLab where I was a developper and administrator for the ImmuneSpace portal. Additionaly, I handled data analysis as well as the development and maintainenance of R package for various projects.
In 2011, I completed my master degree in Bioinformatics and Biostatistics at Université Paris-Sud. My training ended with a six months internship in the Department of Biochemistry and Biophysics, at Stockholm University under the supervision of Prof. Arne Elofsson.